
In recent years, the rapid advancement of techniques such as super-resolution fluorescence imaging has provided unprecedented opportunities for the ultra-high spatiotemporal resolution visualization and biological functional research of target proteins. The high contrast labeling of target proteins is a critical prerequisite for imaging. Due to their unique photophysical properties, small size, and high photostability, small organic molecules have become the most commonly used reporter molecules in protein imaging. However, traditional small organic dyes still face challenges in achieving high contrast labeling of live-cell proteins in biological imaging and sensing, primarily due to insufficient cell permeability and high background signals.
Over the past decade, a new generation of fluorescence probes based on rhodamine structure has been developed, characterized by protein-responsive properties and rapid transmembrane capabilities. These characteristics mainly arise from the dynamic equilibrium between two distinct structures of the dye: a spirolactone structure (closed-ring) that is non-fluorescent in its free state but possesses high transmembrane capability, and a zwitterionic structure (open-ring) that exhibits strong fluorescence upon binding to proteins (Figure 1).
Recently, Dr. Lu Wang, from School of Pharmacy, Fudan University, provides a comprehensive overview of the mechanistic insights and design strategies behind these protein-responsive, rapidly transmembrane rhodamine probes, along with their applications in biological imaging and biosensing.The review ‘Fluorogenic and Cell-Permeable Rhodamine Dyes for High-Contrast Live-Cell Protein Labeling in Bioimaging and Biosensing’ was published on Angew Chem Int Ed.
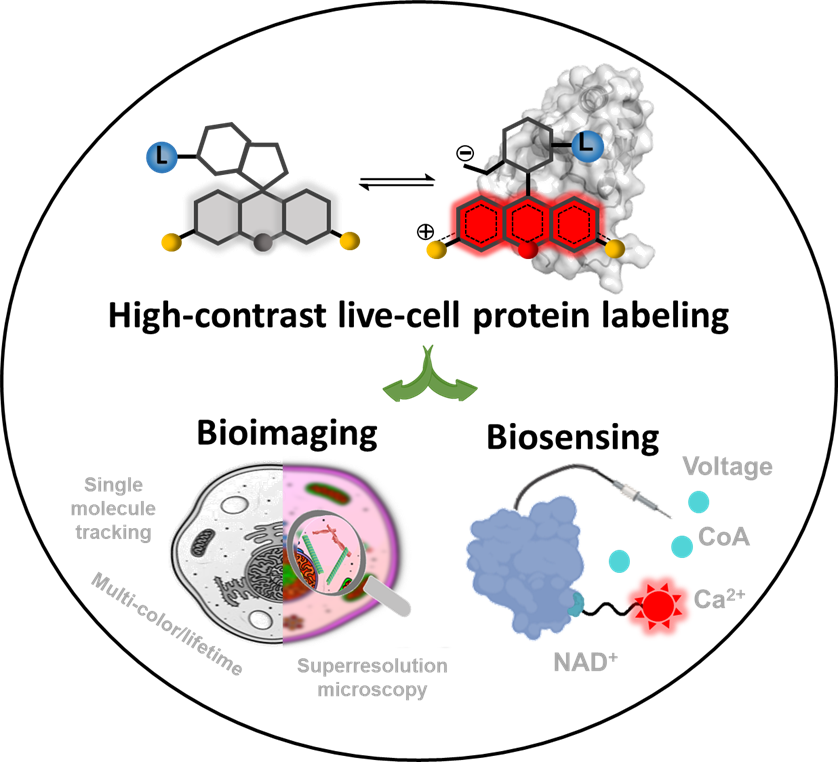
Figure 1 Schematic illustration that fluorogenic and cell-permeable rhodamine probes enable high signal-to-noise ratio labeling of target proteins in live cells, facilitating various biological imaging and biosensing.
Fluorescence probes based on novel rhodamine dyes have achieved high signal-to-noise ratio labeling of target proteins in live cells, including protein tags (such as SNAP tag, Halo Tag, CLIP Tag, and TMP tag), cytoskeletons (such as actin and microtubules), nucleic acids (such as DNA and RNA), organelles (such as lysosomes, mitochondria, Golgi apparatus, and endoplasmic reticulum), and drug targets (such as folate receptors and β-secretase 1). These probes have become the most widely used tools in current live-cell super-resolution imaging, providing unprecedented opportunities for observing cellular structures and studying protein functions. Moreover, in the field of biosensing, the new generation of rhodamine probes has greatly facilitated the construction of chemical-genetic biosensors. These innovative chemical-genetic biosensors combine the excellent photostability and photophysical properties of organic fluorophores with the precise targeting and selectivity of proteins, enabling successful monitoring of metabolites, signaling molecules, and voltage dynamics within live cells. With the continuous development of fluorogenic and cell-permeable dyes, it is anticipated that more effective tools for biological imaging and biosensing will be prepared, significantly advancing the field of biomedical research and development.
The co-first authors of this paper are Dongjuan Si, from School of Pharmacy, Fudan University, and Quanlin Li, from Zhongshan Hospital. Dr. Jingye Zhang and Dr. Lu Wang are the corresponding authors of the paper. Yifan Bao also contributed to the writing of the paper. This work received support from the National Natural Science Foundation of China through its General and Youth Programs, the Shanghai Basic Research Program, the Zhangjiang mRNA Innovation and Translation Center, and other projects.
Article links: https://onlinelibrary.wiley.com/doi/10.1002/anie.202307641