
Recently, Professor Jianxin Wang’s group from the School of Pharmacy, Fudan University, in collaboration with Researcher Shuangjia Zheng from Shanghai Jiao Tong University and others, published a study entitled “Geometric-aware deep learning enables discovery of bifunctional ligand-based liposomes for tumor targeting therapy” in Nano Today. This study combines geometric-aware deep learning technology with wet-lab validation to successfully screen novel natural products with dual functionalities of lipid membrane regulator (LMR) and glucose transporter 1 (Glut1) ligands from over 300,000 natural compounds. Based on these bifunctional natural products, the researchers developed bifunctional natural product-based liposomes for tumor targeting therapy, paving the way for a new paradigm in intelligent drug delivery system design.
Traditional liposomes have limited tumor-targeting capabilities, which compromise their clinical efficacy. Although ligand-modified liposomes have shown potential in enhancing tumor-targeting efficiency, their complex manufacturing processes hinder clinical translation. Previous studies by the Prof. Wang’s team revealed that certain natural products possess both lipid membrane regulation (LMR) and tumor-targeting functions. This discovery suggests that replacing cholesterol in liposomes with these natural products could achieve tumor targeting without the need for ligand synthesis. However, with over 300,000 natural products available, traditional trial-and-error screening for such bifunctional natural products is time-consuming and labor-intensive.
To address this challenge, this study integrates artificial intelligence deep learning models with wet-lab validation to rapidly identify novel bifunctional natural products. By dataset collecting, geometric-aware deep learning model constructing, performance predicting, and experimental validation conducting, six bifunctional natural products were efficiently screened from over 300,000 candidates. Among them, liposomes constructed with Ilexgenin A (Ile) demonstrated the best tumor-targeting capability and antitumor efficacy, confirming the immense potential of deep learning technology in designing intelligent targeted drug delivery systems.
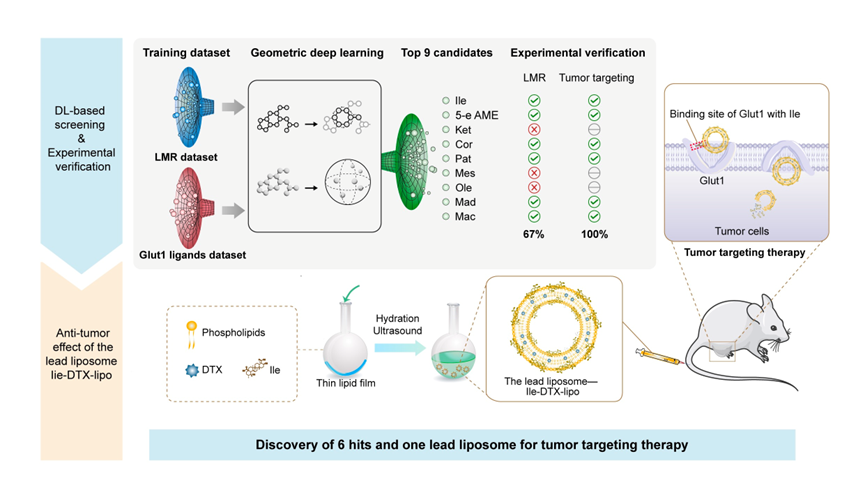
Workflow of the DL-guided approach for discovery of natural product-based liposomes
As shown in Figure 1, the research team developed a Geometric-aware Message Passing Neural Network (GMPNN) that infers three-dimensional conformational information from two-dimensional molecular structures through a contrastive learning strategy. This approach overcomes the limitations of traditional deep learning models, which rely heavily on large datasets and have insufficient geometric feature modeling.
After training on collected datasets of lipid membrane regulators (LMR) and glucose transporter 1 (Glut1) ligands, the model demonstrated outstanding performance in LMR prediction and Glut1 binding activity prediction tasks, significantly outperforming traditional methods such as Support Vector Machine (SVM) and Random Forest (RF).
Using this model, the team performed virtual screening of over 300,000 molecules from the Dictionary of Natural Products (DNP). By combining structural diversity and similarity analysis, nine candidate molecules were identified: Madecassic acid (Mad), Betulin, Ilexgenin A (Ile), Corosolic acid (Cor), Macranthoidin A (Mac), 5-Ene-24-Oic Acid-3β-Methyl Ester (5-e AME), Mestanolone (Mes), Oleandrin (Ole), and 4-Pregnen-3,11,20-Trione (Ket).
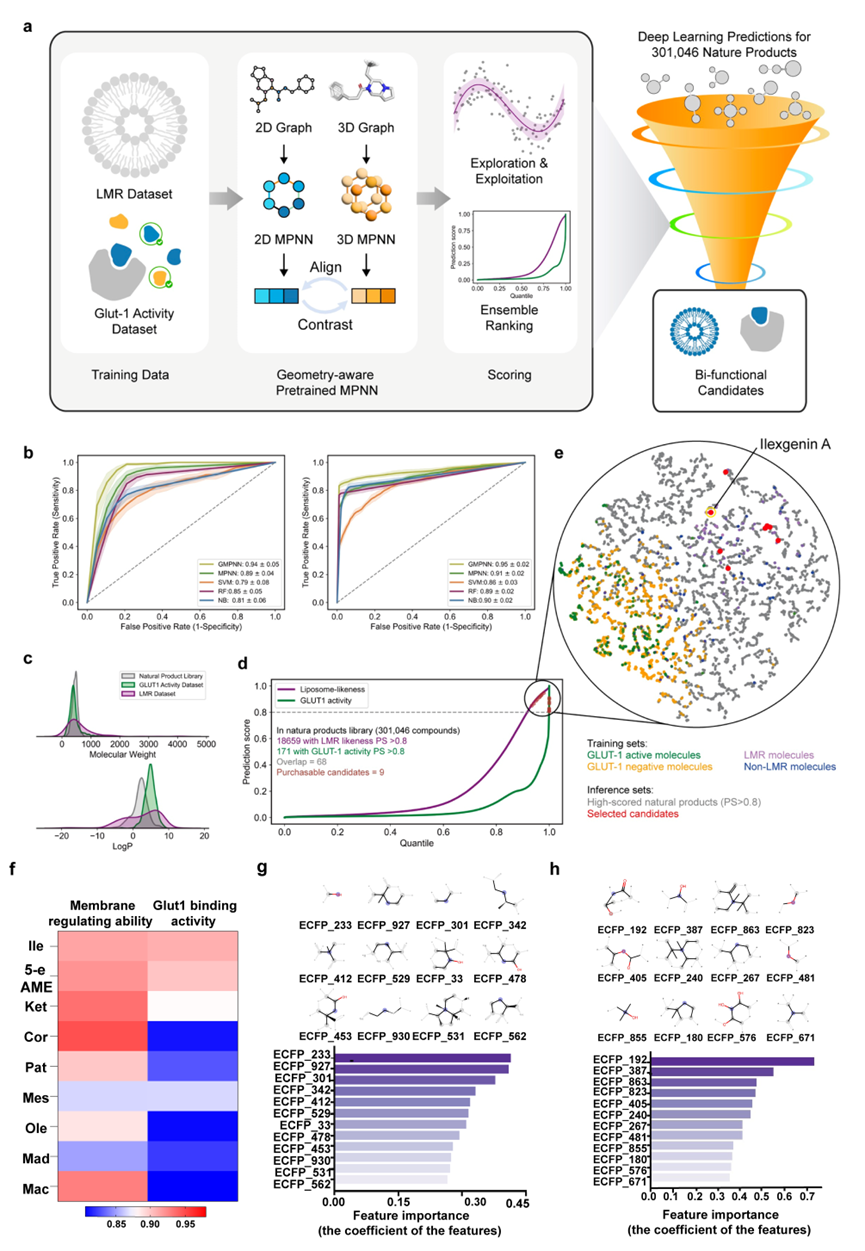
Figure 1. Construction of the GMPNN Algorithm Model and Identification of Bifunctional Candidates
Wet-lab validation results demonstrated that among the nine GMPNN-selected candidates, six (excluding Mes, Ket, and Ole) successfully replaced cholesterol as LMRs and be prepared as liposomes with uniform particle size (approximately 100 nm) and good stability (Figure 2).
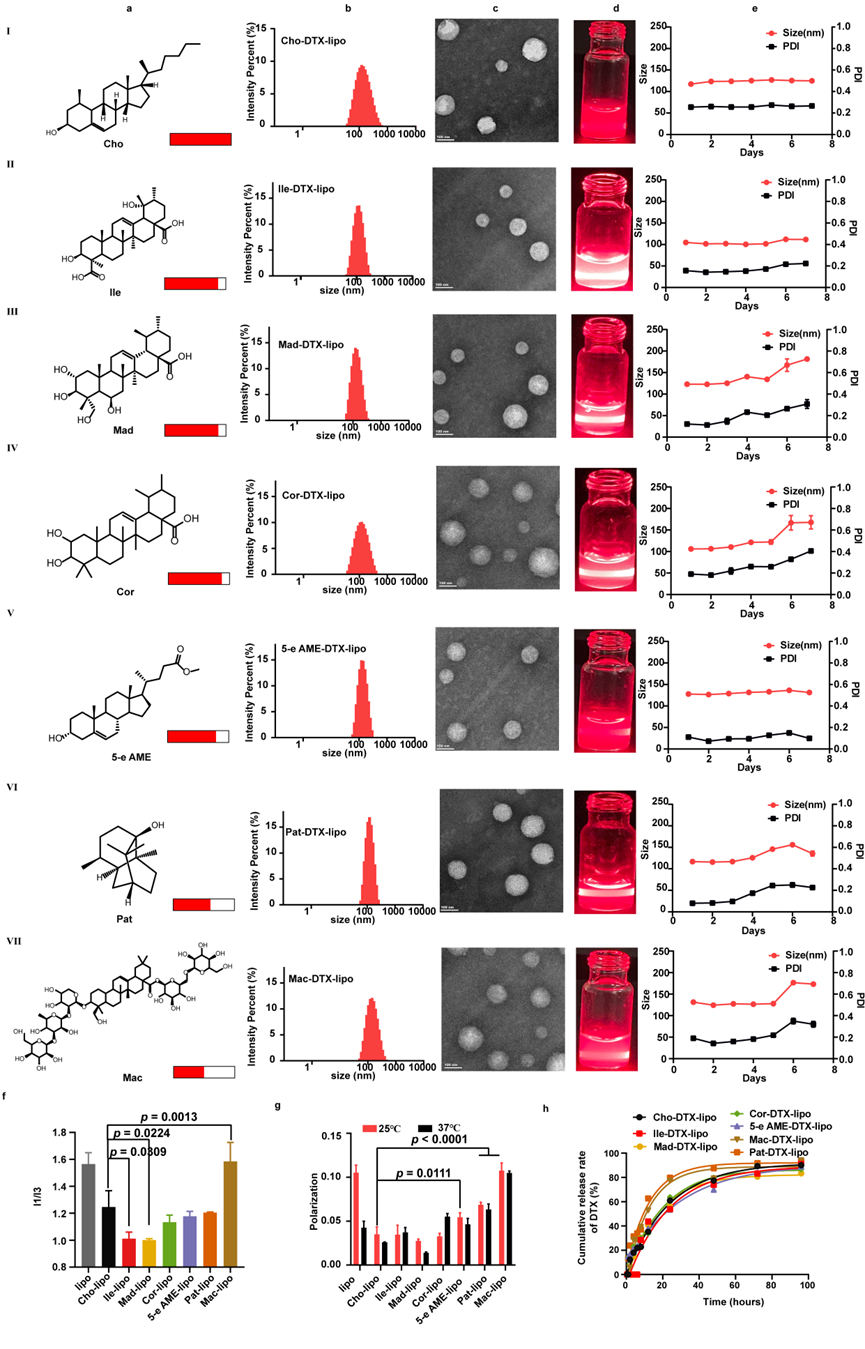
Figure 2. Wet-Lab Validation of LMR Activities for GMPNN-Selected Candidates
The above LMR-active candidates all exhibited significant Glut1-mediated tumor-targeting capabilities, with their liposome formulations demonstrating superior tumor-targeting performance compared to conventional cholesterol-based liposomes. Furthermore, Ilexgenin A liposomes (Ile-lipo) achieved the highest tumor-targeting efficiency, even significantly outperforming the previously discovered ginsenoside-based liposomes by the research team (Figure 3).
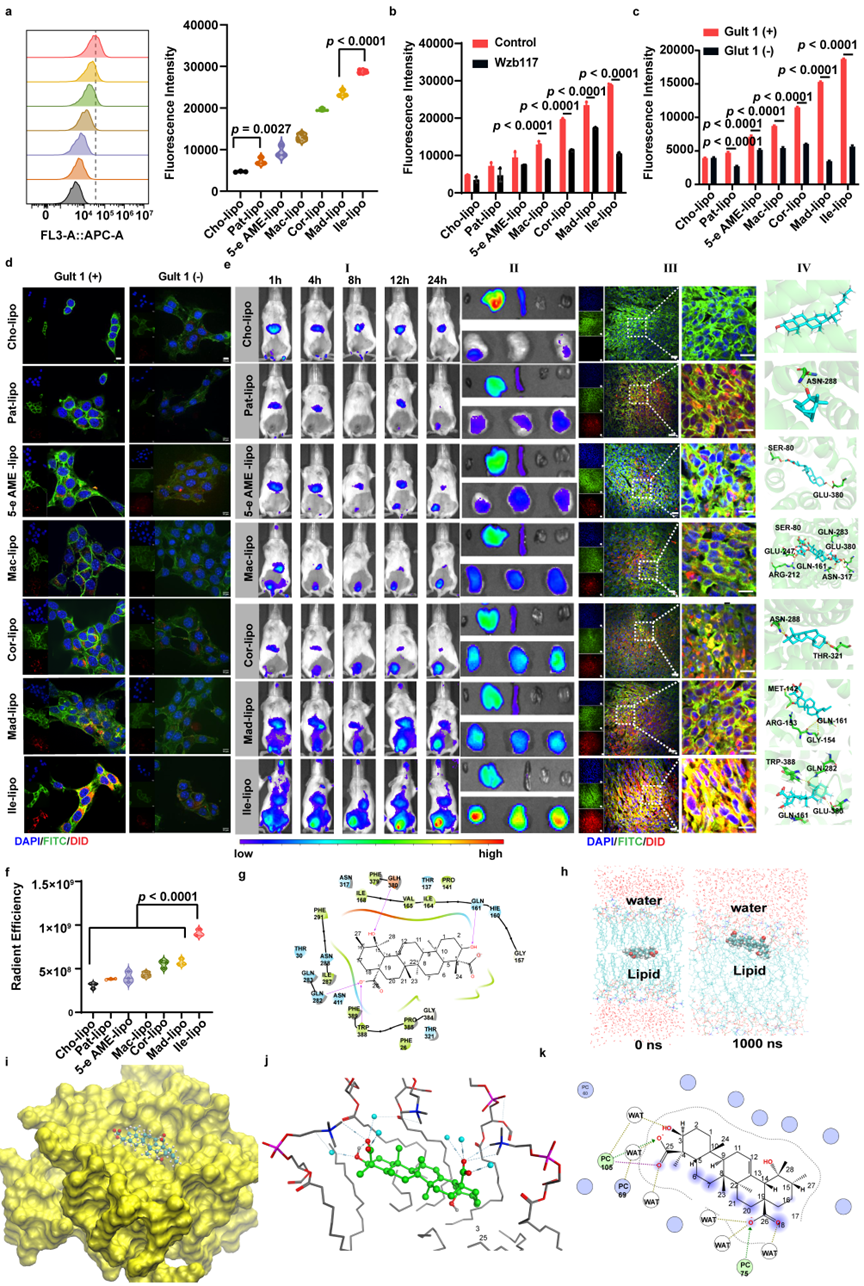
Figure 3. Wet-Lab Validation of Tumor-Targeting Activity of the Liposomes
Finally, Ilexgenin A based liposomes loaded with the chemotherapy drug docetaxel (Ile-DTX-lipo) demonstrated potent antitumor activity due to their excellent tumor-targeting ability (Figure 4), making them an effective tumor-targeted drug delivery strategy.
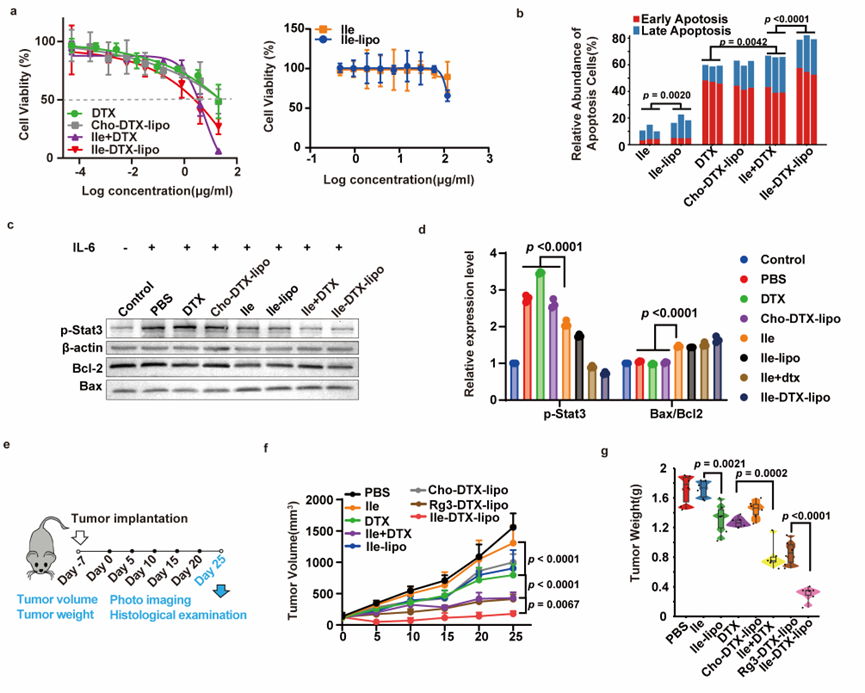
Figure 4. Anti-tumor Effect of Ile-DTX-lipo
This study innovatively applied geometric-aware deep learning for the efficient discovery of bifunctional ligands, overcoming the limitations of traditional screening methods, with success rates increasing to 67% (for LMR function prediction) and 100% (for Glut1 binding function prediction). Ilexgenin A based liposomes, with their simple preparation process, excellent targeting ability, and immune-regulatory functions, show significant clinical translation potential. In the future, this strategy can be extended to the development of intelligent delivery systems targeting other disease sites, providing new tools for precision medicine.
Postdoctoral researcher Jiaxuan Xia and master's student Zicheng Gan from the School of Pharmacy, Fudan University, are co-first authors of the paper. Chengtao Li from Xingyao Technology Co., Ltd., Shuangjia Zheng from the Pu Yuan Institute of Future Technologies, Shanghai Jiao Tong University, and Professor Jianxin Wang from the School of Pharmacy, Fudan University, are co-corresponding authors. The research was supported by the National Natural Science Foundation of China (82374296, 82074277) and other projects.
Paper link: https://doi.org/10.1016/j.nantod.2025.102668